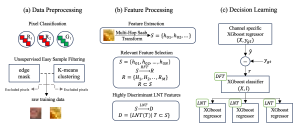
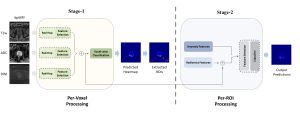
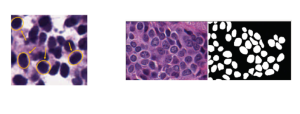
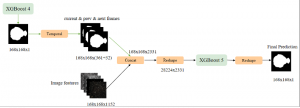
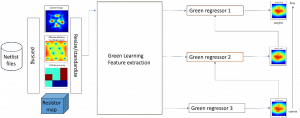
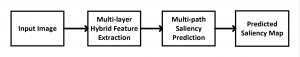
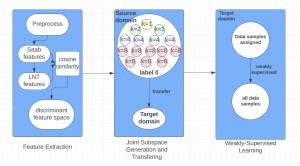
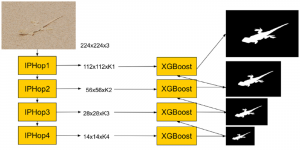
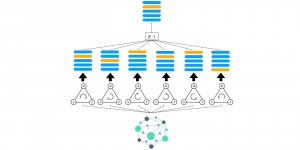
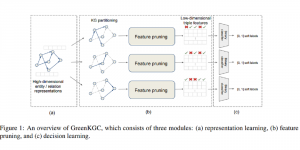
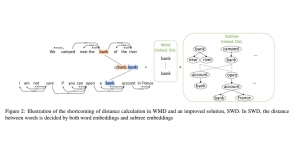
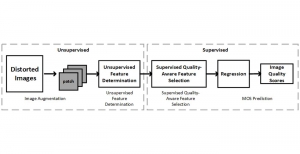
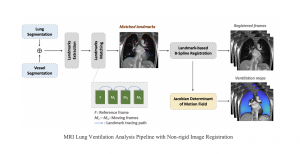
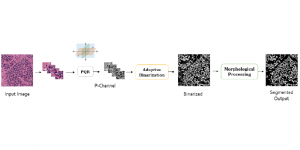
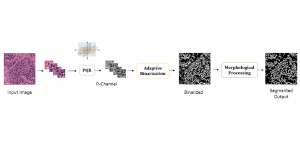
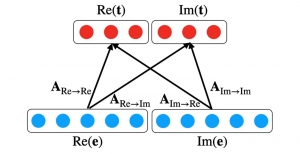
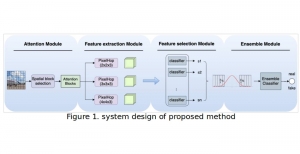
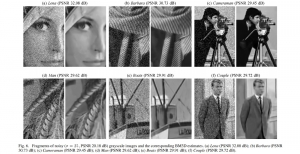
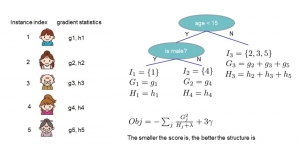
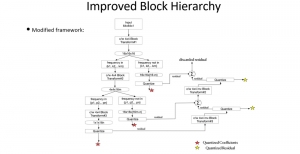
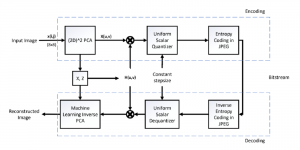
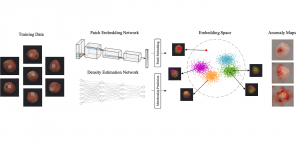
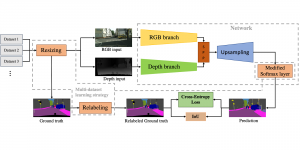
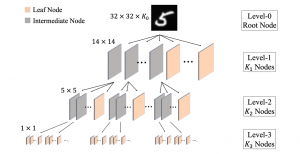
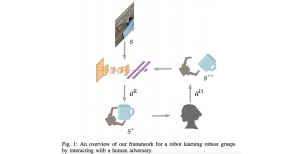
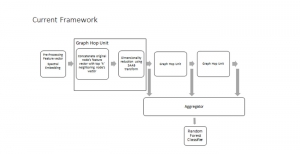
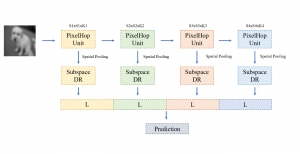
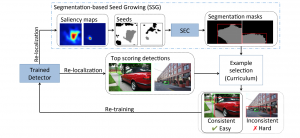
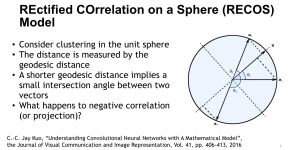
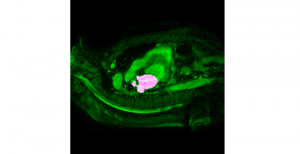
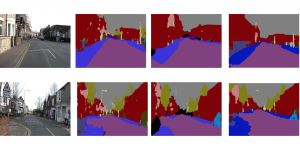
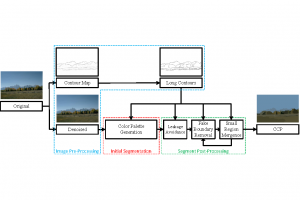
Biomedical image segmentation is a technique for automatic detection of organ boundaries within an image, used to obtain diagnostic insights in the field of medicine. Segmentation results are used in applications involving organ measurements, cell detection and blood flow simulations [1].
Zebrafish is a vertebrate that have similar organs and tissues as humans, making it a valuable model for studying human genetics and disease [2]. In this project, we use the zebrafish organs dataset for the analysis and evaluation of medical image segmentation methodologies.
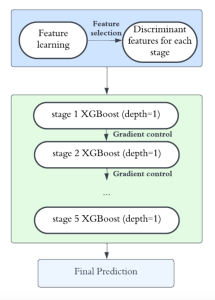
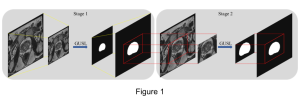
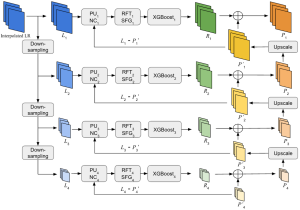
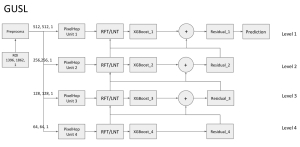
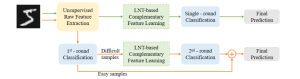
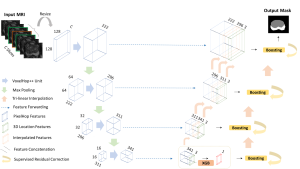
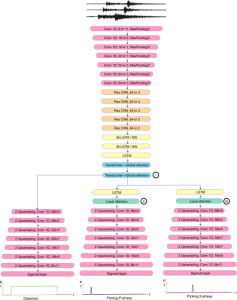
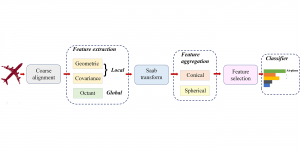
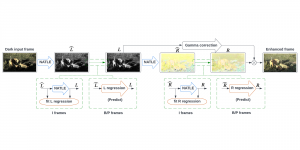
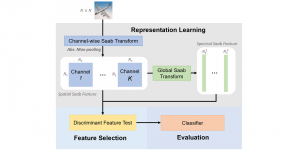
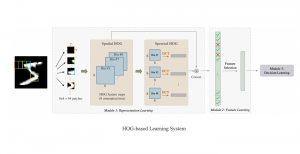
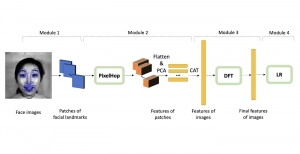
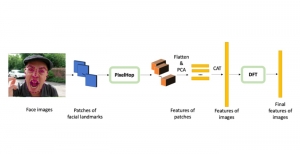
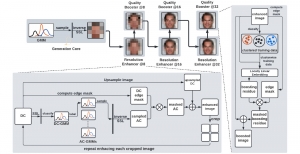
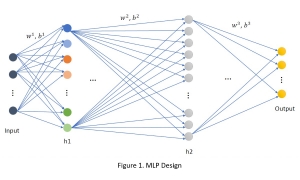
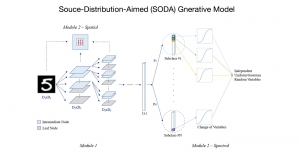
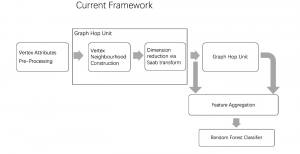
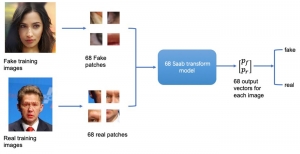
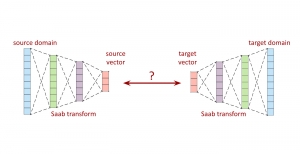
Presently, we have trained a CNN based segmentation model using RefineNet [3] and a Saak [4] based model on the zebrafish data. Our goal is to compare and analyze the performance of CNN with Saak segmentation model. In future, we aim to improve these results by adopting content adaptive Saak with clustering techniques and statistical analysis algorithms.
-By Shilpashree Rao and Ruiyuan Lin
Reference:
[1]”Medical Image Segmentation”. [Online]. Available: https://www5.cs.fau.de/research/groups/medical-image-segmentation. [Accessed: 01- Apr- 2018].
[2]”Why use the zebrafish in research?”, 2018. [Online]. Available: https://www.yourgenome.org/facts/why-use-the-zebrafish-in-research. [Accessed: 01- Apr- 2018].
[3] G. Lin, A. Milan, C. Shen and I. Reid, “RefineNet: Multi-Path Refinement Networks for High-Resolution Semantic Segmentation,” in IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2017.
[4] C.-C. J. Kuo and Y. Chen, “On data-driven Saak transform,” Journal of Visual Communication and Image Representation, vol. 50, pp. 237–246, 2018.
Image credits:
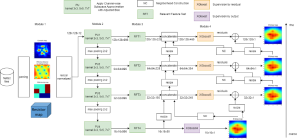
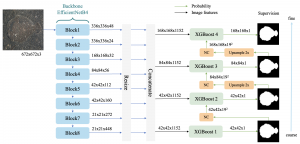
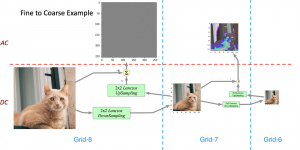
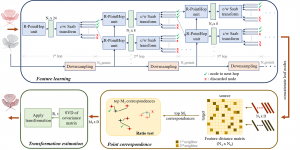
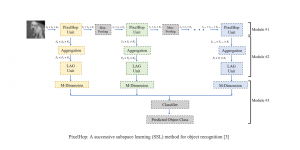
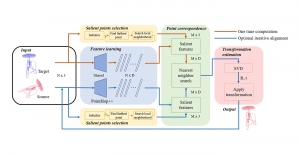
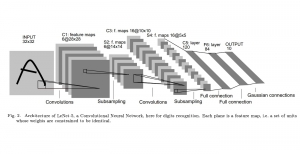
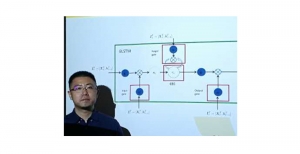
Image showing the architecture of RefineNet is from [3].
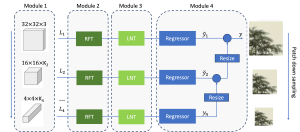
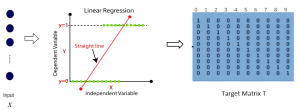
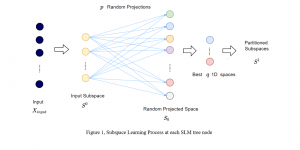
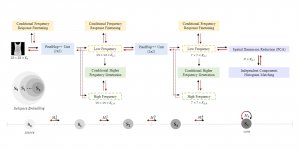
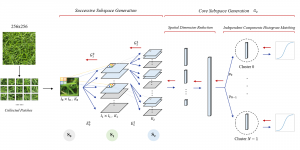
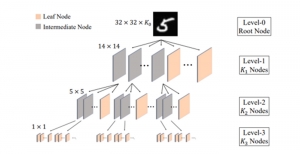
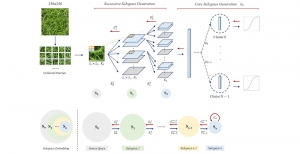
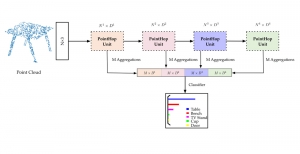
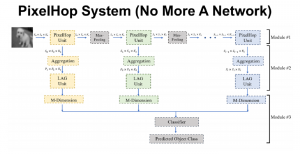
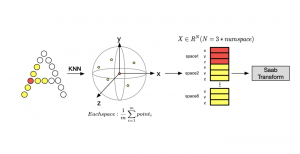
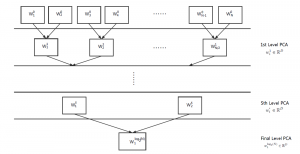
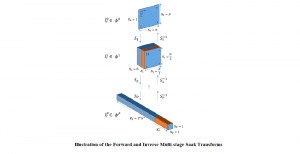
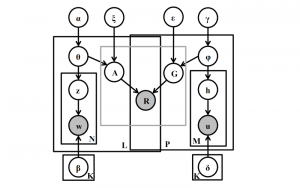
Image showing the architecture of multi-stage Saak is from [4].