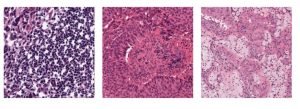
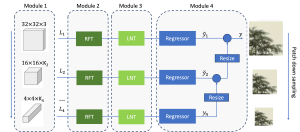
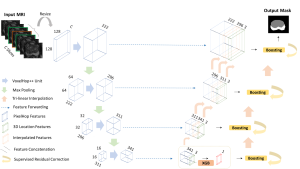
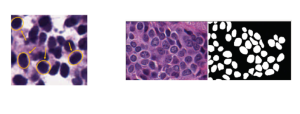
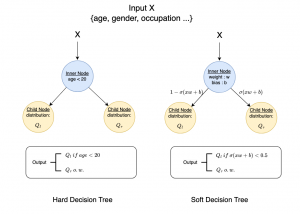
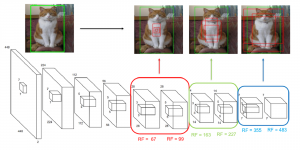
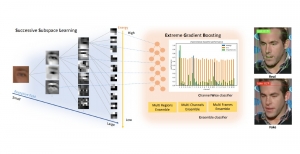
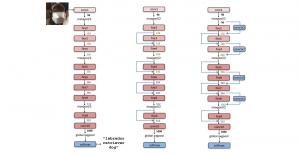
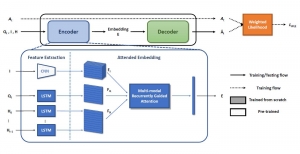
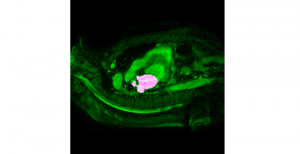
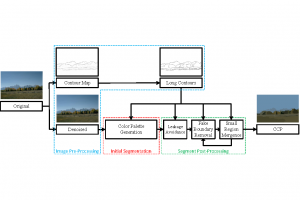
Nuclei segmentation is a consequential task in biological image analysis, helping in the reading process of histology images. Different attributes, such as shape, population, cluster formation and density play a significant role in clinical practice for cancer diagnosis and its aggressiveness level assessment. Given that the annotation of this data is carried out by expertized pathologists who reportedly [2] need to spend on average 120-150 hours to annotate 50 image patches (about 12M pixels), one can realize that annotated data are in scarcity. That is a big impediment for supervised methods, particularly for DL-based solutions that need massive annotated data to learn generalizable representations. Moreover, the annotations have a high inter-observer variation which is subject to the experience of the annotator [1]. On top of that, nuclei color and texture variations across images from different laboratories and multiple organs further widen the gap between train and test domains.
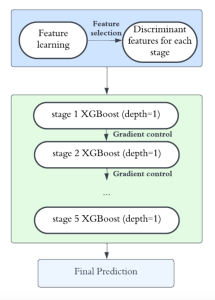
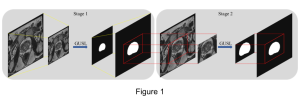
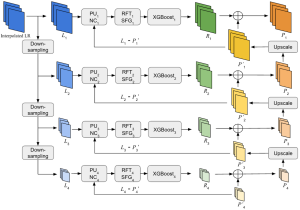
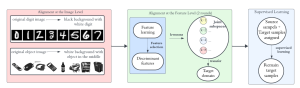
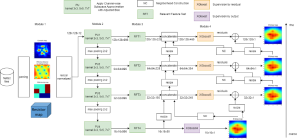
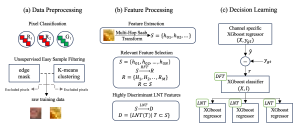
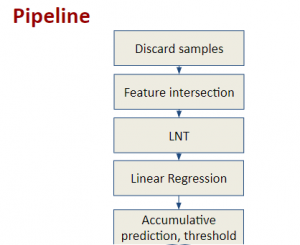
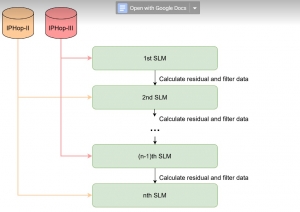
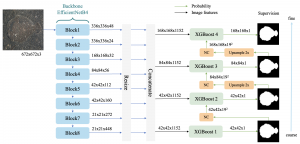
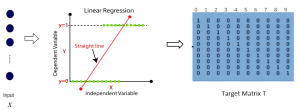
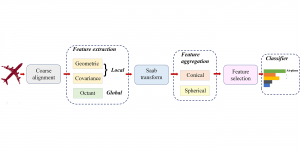
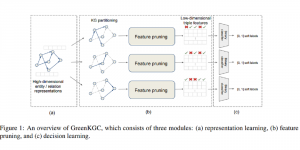
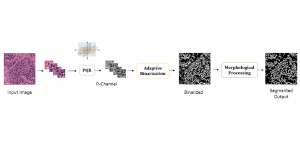
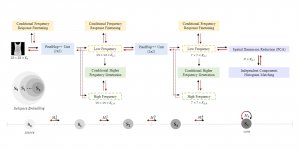
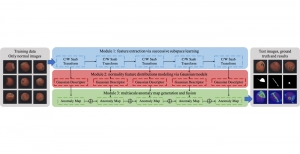
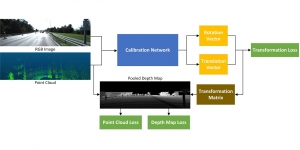
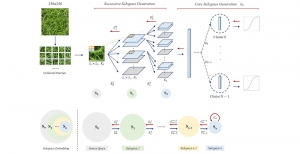
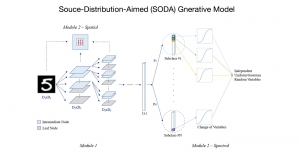
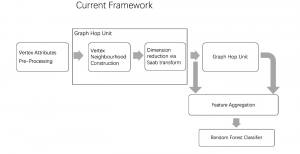
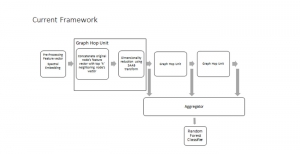
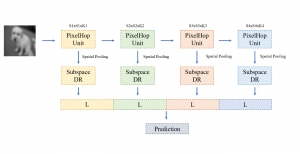
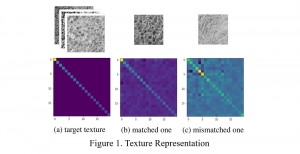
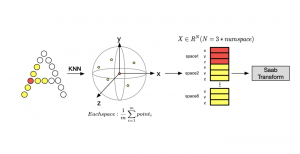
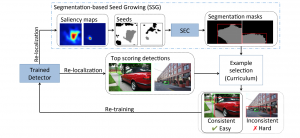
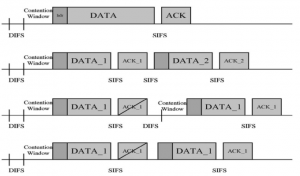
Given the aforementioned limitations, a natural way to solve the problem is to pursue an unsupervised line of research. Also, given the limited number of annotated data, our proposed method decouples from the DL paradigm and utilizes conceptually simpler techniques that make the pipeline more transparent in terms of segmentation decision making. It is mainly based on prior knowledge about the nuclei segmentation problem. CBM [3] pipeline starts out with a data-driven Color (C) transform, to highlight the nuclei cell regions over the background, followed by an adaptive Binarization (B) process built on the bi-modal assumption in each local region. That process is being run in a patch-wise manner, to leverage the local distribution assumptions between background and foreground. The final part of the pipeline uses Morphological (M) transformations that refines the segmented output based on certain priors about the cell size and shape.
The proposed methodology outperforms other unsupervised DL methods by large margins and has a very competitive standing among the supervised DL methods in the MoNuSeg public dataset, maintaining a low complexity because of its parameter-free design. As a future direction, we plan to solve the over-segmentation issue, that is common in unsupervised thresholding methods, by incorporating a second stage for attribute learning and nuclei shape refinement, based on the labels from CBM (1st stage). Hence, by creating some self-supervision signal to be fed in the second stage, we may reduce the false positive number of detected nuclei instances.
— By Vasileios Magoulianitis
[1] H. Irshad, L. Montaser-Kouhsari, G. Waltz, O. Bucur, J. Nowak, F. Dong, N.W. Knoblauch, and A. H. Beck, “Crowdsourcing image annotation for nucleus detection and segmentation in computational pathology: evaluating experts, automated methods, and the crowd,” in Pacific symposium on biocomputing Co-chairs. World Scientific, 2014, pp. 294–305.
[2] L. Hou, A. Agarwal, D. Samaras, T. M. Kurc, R. R. Gupta, and J. H. Saltz, “Robust histopathology image analysis: To label or to synthesize?” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2019, pp. 8533–8542.
[3] Magoulianitis, V., Han, P., Yang, Y., & Kuo, C. C. J. (2021). Unsupervised Data-Driven Nuclei Segmentation For Histology Images. arXiv preprint arXiv:2110.07147.