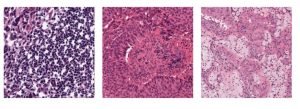
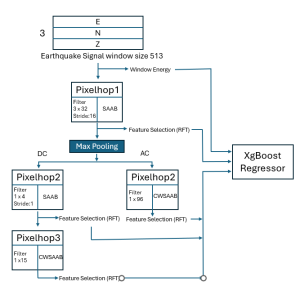
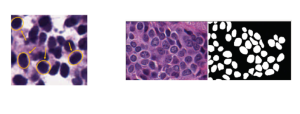
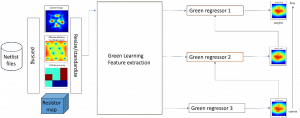
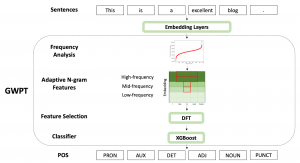
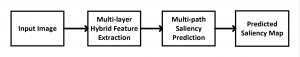
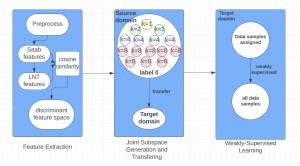
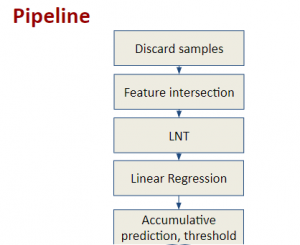
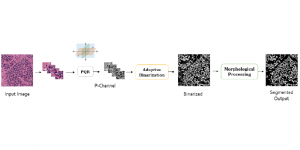
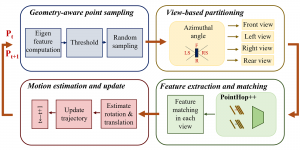
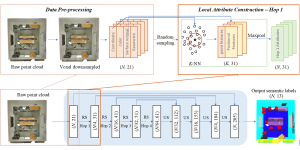
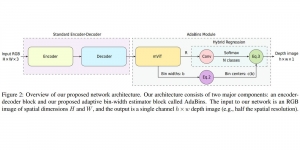
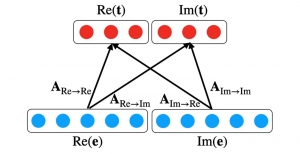
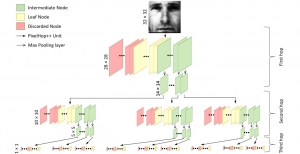
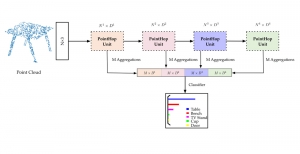
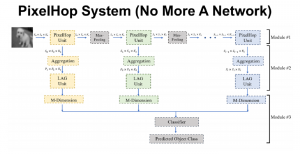
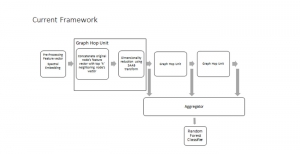
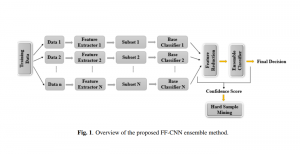
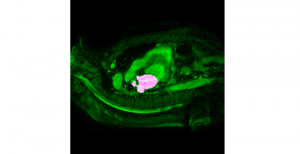
Nuclei segmentation has been extensively used in biological image analysis for reading histology images from microscopes. The population of nuclei, their shape and density play a significant role in clinical practice for cancer diagnosis and its aggressiveness assessment. The reading and annotation of those images is a fairly laborious and time consuming task, being carried out only from expertised pathologists. As such, the computer aided automation of this process is of high significance, since it reduces the physicians’ work load and provides a more objective segmentation output. Yet, different challenges such as color and intensity variations that result from images of different organs and acquisition settings, hinder the performance improvement of the algorithms.
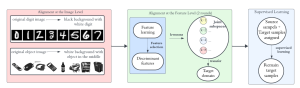
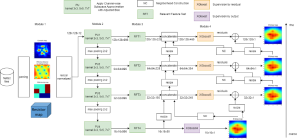
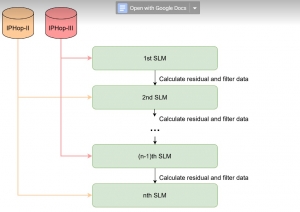
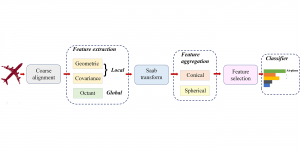
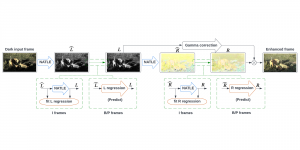
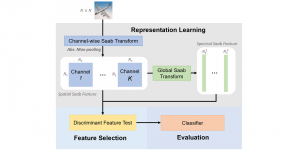
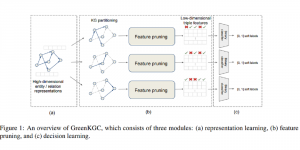
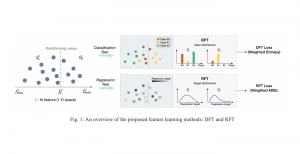
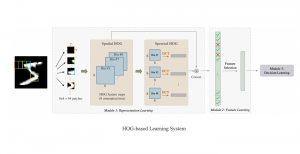
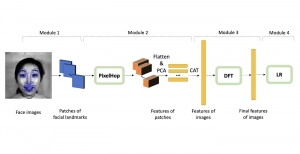
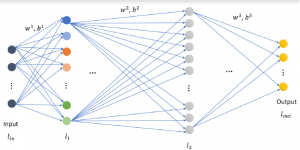
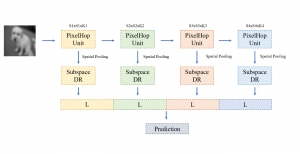
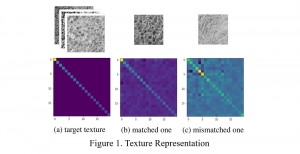
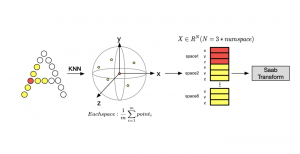
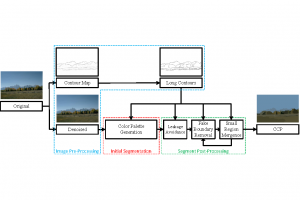
In past years, both supervised and unsupervised solutions have been proposed to tackle those challenges. Most of the recent approaches [1] adopt deep learning (DL) networks, to directly learn from pathologists’ annotated segmentation masks. However, most of the available datasets have very few samples, relatively to the DL requirements, and also their annotations have reportedly a low inter-annotator rate of agreement. Therefore, it is quite challenging for DL models to generalize well to unseen images from multiple organs. At this point, the motivation behind using an unsupervised method should be obvious. We propose a data driven and parameter free methodology [2], named CBM, for the nuclei segmentation task that requires no training labels. The pipeline begins with a data-driven Color (C) transform, to highlight the nuclei cell regions over the background. Then, a data-driven Binarization (B) process is built on the bi-modal assumption that each local region has two histogram peaks, one corresponding to the background and one to the nuclei areas. We use a locally adaptive threshold to binarize each histology image in a patch-based manner. The final part of our pipeline uses Morphological (M) transformations that refines the segmented output based on certain priors about the cell size and shape. Moreover, it is efficient in splitting the falsely connected nuclei cells, by adopting a technique based on the nuclei convexity prior.
The proposed methodology outperforms other unsupervised methods and has a very competitive standing among the supervised DL methods in the MoNuSeg public dataset, having the additional merit that it requires no annotations. Also, it is a parameter free and computationally inexpensive pipeline, being transparent to physicians, contrary to the DL models output that their decisions are hard to be explained. An interesting direction to be explored in the future is self or weakly supervision to further improve the performance, while keeping the labels requirement as low as possible.
— By Vasileios Magoulianitis
[1] Y. Zhou, et. al. “Cia-net: Robust nuclei instance segmentation with contour-aware information aggregation,” in International Conference on Information Processing in Medical Imaging. Springer, 2019, pp. 682–693.
[2] Magoulianitis, V., Han, P., Yang, Y., & Kuo, C. C. J. (2021). Unsupervised Data-Driven Nuclei Segmentation For Histology Images. arXiv preprint arXiv:2110.07147.