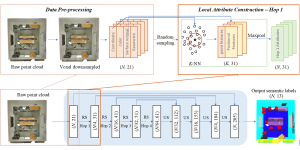
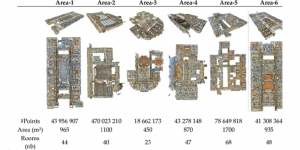
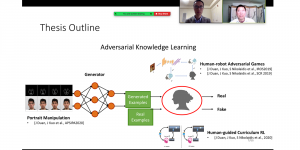
Research in healthcare systems has been mainly focused on automating several tasks in the clinical pipeline, aiming at enhancing or expediting physician’s diagnosis. Prostate Cancer (PCa) is widely known as one of the most frequently occurring cancer in diagnosis in men. If early diagnosed, the mortality rate is almost zero. Yet, should it goes under the radar -in the metastasis stage- that rate plummets at 31% [1]. For diagnosis, after a high level of Prostatic Specific Antigen (PSA), patients are recommended to undergo an MRI screening. Those patients with suspiciously looking lesions on the prostate gland will eventually undergo biopsy. The histology gives the definite answer whether a patient suffers from cancer. Nevertheless, it is observed in real practice that urologists tend to over-diagnose patients with csPCa, thus increasing the number of unnecessary biopsies. As such, this increases the diagnostic costs and patient discomfort.
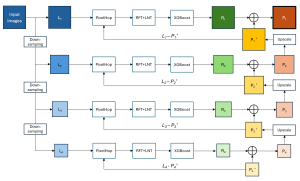
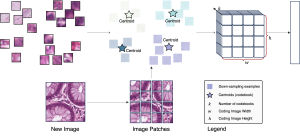
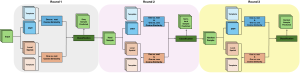
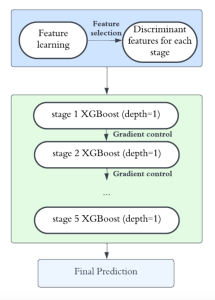
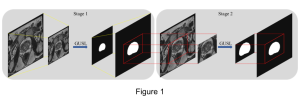
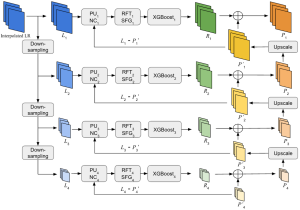
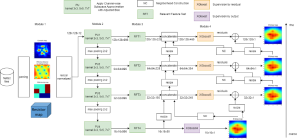
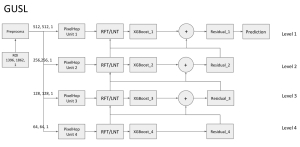
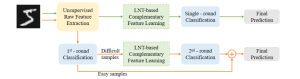
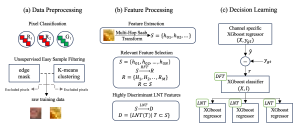
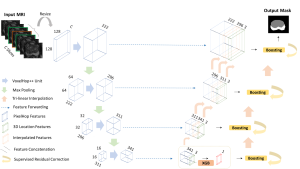
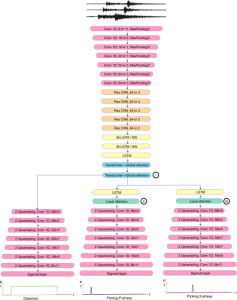
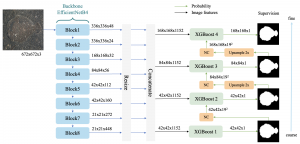
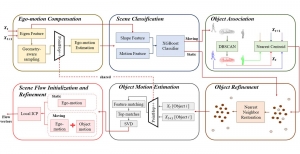
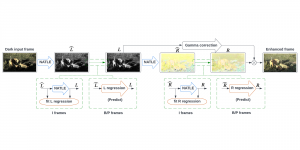
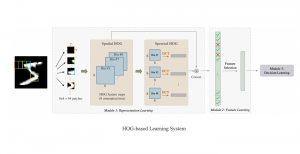
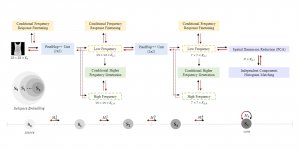
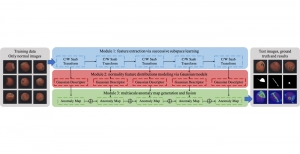
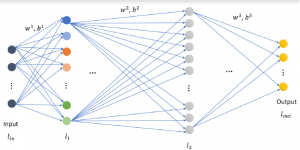
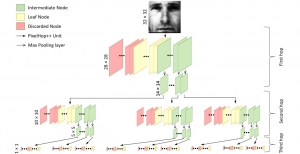
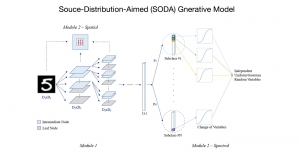
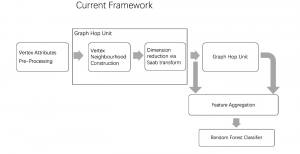
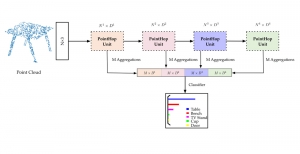
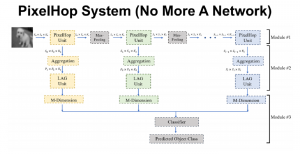
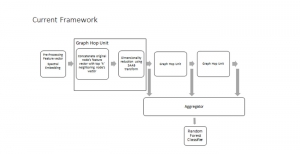
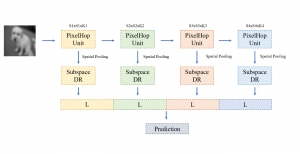
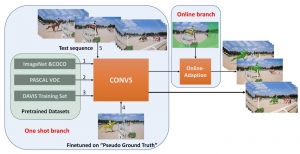
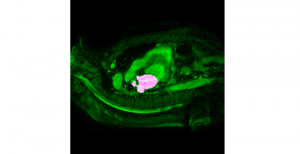
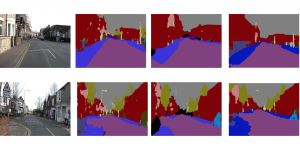
Computer vision empowered with AI has shown promising results in the last decade. Computer-Aided Diagnosis (CAD) tools benefit from AI’s rapid evolution and many works have been proposed to automatically perform lesion detection and segmentation. Even though the Deep Learning (DL) paradigm is ubiquitous in modern AI, medical applications require more transparency behind feature extraction and thereby DL is often deemed as a “black-box” from physicians. Our proposed pipeline, PCa-RadHop [2], employs a novel and linear module for data-driven feature extraction and hence the decision making becomes more interpretable. PCa-RadHop receives three different modalities as input from the MRI-scanner (i.e. T2w, ADC, DWI), pertinent to PCa diagnosis. It consists of two stages. The first stage calculates a probability map about csPCa presence in a voxel-wise manner, while the second stage is meant to reduce the false positive rate on that heatmap by employing handcrafted radiomics features. Finally, it yields the Regions of Interest with a corresponding probability of being csPCa.
PCa-RadHop achieved an AUC score of 0.807 at the PI-CAI challenge [3], in detecting csPCa on a testing cohort of 1,000 patients. Moreover, the model size and complexity are orders of magnitude lower compared to other state-of-the-art DL-based solutions.
[1] Siegel, R.L., Miller, K.D., Wagle, N.S., Jemal, A., 2023. Cancer statistics,
2023. CA: a cancer journal for clinicians 73, 17–48
[2] Magoulianitis, V., Yang, J., Yang, Y., Xue, J., Kaneko, M., Cacciamani, G., … & Nikias, C. (2024). PCa-RadHop: A transparent and lightweight feed-forward method for clinically significant prostate cancer segmentation. Computerized Medical Imaging and Graphics, 102408.
[3] Saha, A., Bosma, J., Twilt, J., van Ginneken, B., Yakar, D., Elschot, M.,
Veltman, J., Fütterer, J., de Rooij, M., et al., 2023. Artificial intelligence
and radiologists at prostate cancer detection in mri—the pi-cai challenge,
in: Medical Imaging with Deep Learning, short paper track.