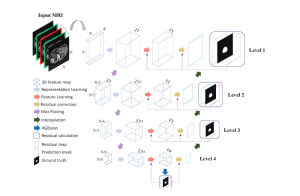
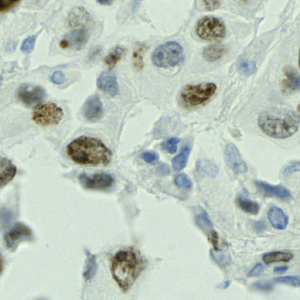
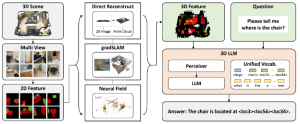
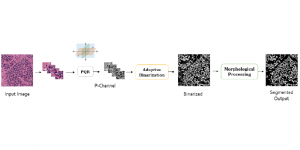
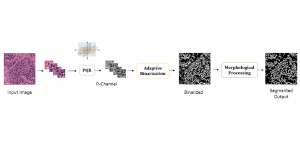
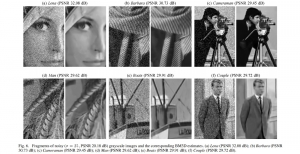
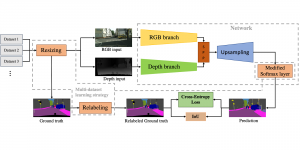
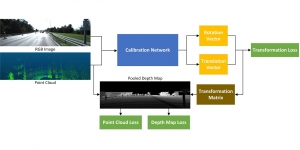
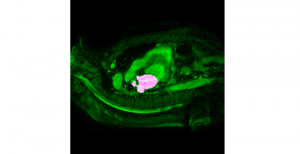
Nuclei segmentation is a fundamental task required to analyze the underlying nuclei structure of an organ of interest. Cancer starts from the cells, and understanding the nuclei shapes, sizes and distribution can provide cues on whether or not a patient has cancer. Further analysis can also help in cancer grading and prognosis. However, studying whole slide images of biopsied tissues requires a large amount of time and effort.
Such monotonous and laborious tasks can be simplified by using AI and ML, and can perhaps improve the accuracy of the detections as well. Some challenges in the nuclei segmentation task include inherent staining variations in the WSI, a wide variety of shapes and sizes in nuclei, and irregular boundaries which make it difficult to track the actual contours. Most of the current research in this area involves deep learning based architectures like the U-Net, R-CNN, and even Vision Transformer. These methods require a large number of training samples and high complexity to achieve generalization among the variations inherent in nuclei from different organs.
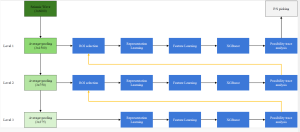
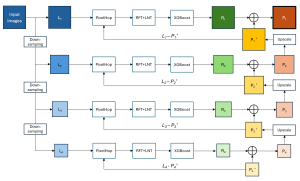
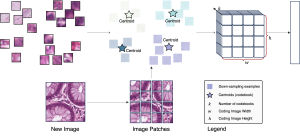
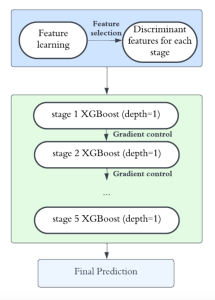
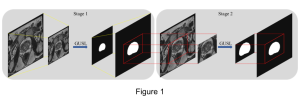
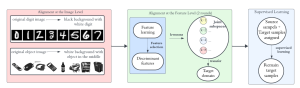
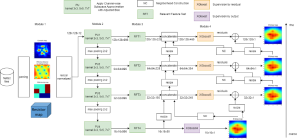
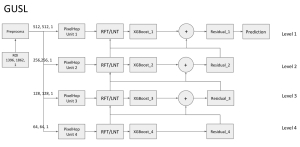
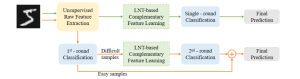
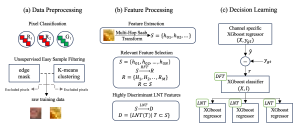
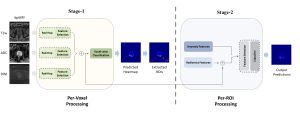
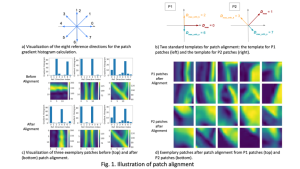
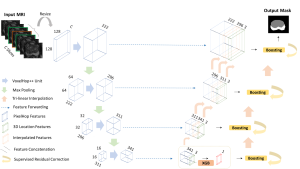
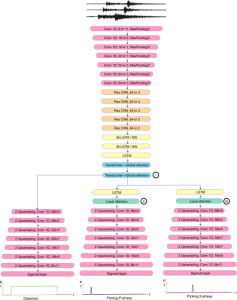
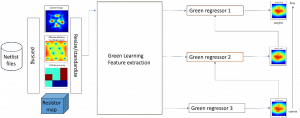
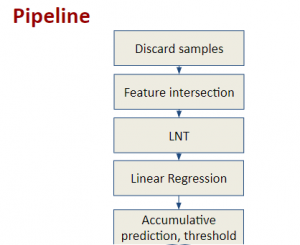
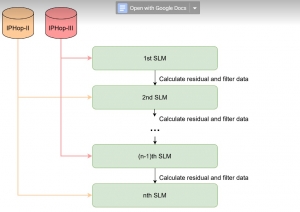
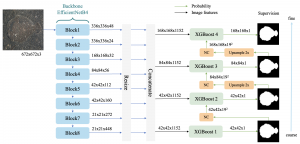
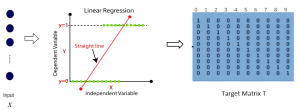
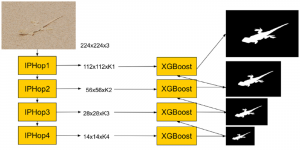
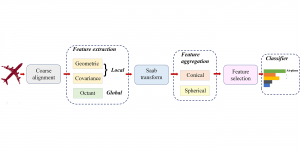
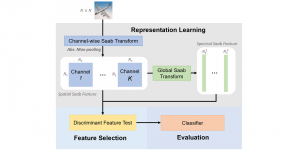
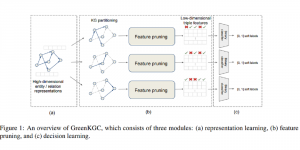
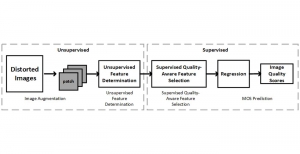
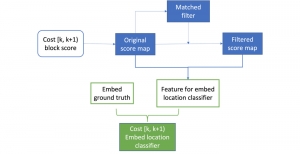
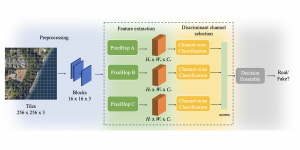
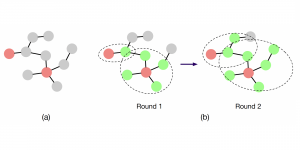
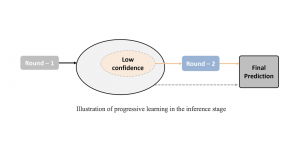
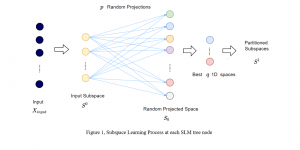
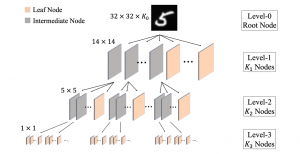
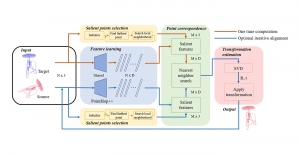
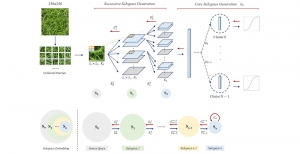
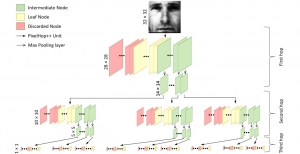
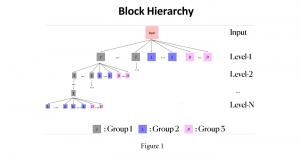
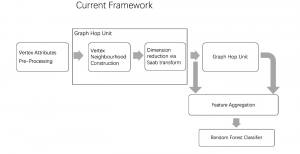
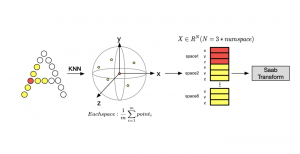
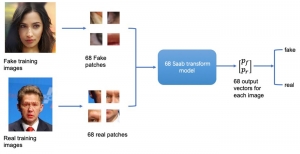
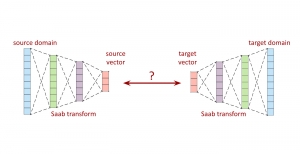
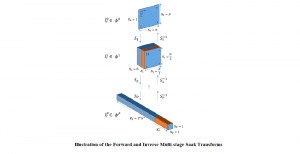
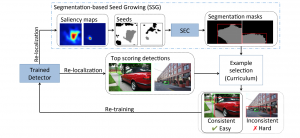
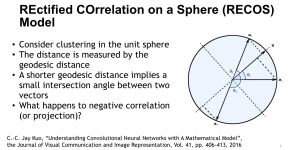
We propose to use a light-weight, interpretable, and simple Green Learning based approach to perform Nuclei Segmentation. Prior work on highly effective Unsupervised Nuclei Instance Segmentation (HUNIS) [1] forms the first stage of our current approach. To further improve HUNIS results, we now focus on the regions where HUNIS requires the help of labels. We divide our current task into two stages: (i) to identify those areas where we need help and (ii) to correct those areas towards their actual class. With the help of Saab Transform, our main task now is to perform feature engineering to identify the ideal features to implement the above two stages.
References:
[1] V. Magoulianitis, Y. Yang, and C.-C. J. Kuo, “HUNIS: High-Performance Unsupervised Nuclei Instance Segmentation,” in 2022 IEEE 14th Image, Video, and Multidimensional Signal Processing Workshop (IVMSP), IEEE, 2022, 1–5.