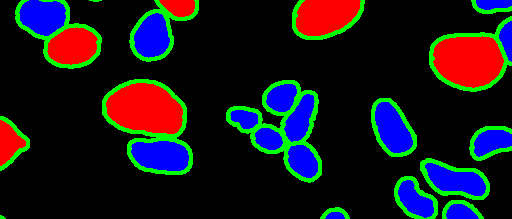
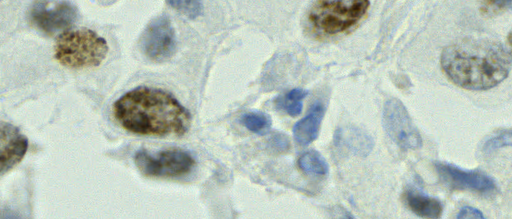
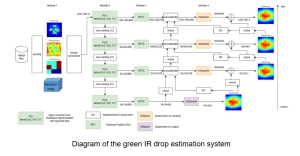
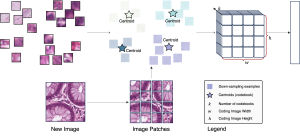
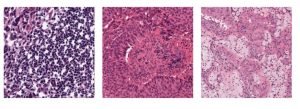
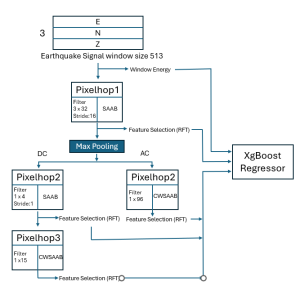
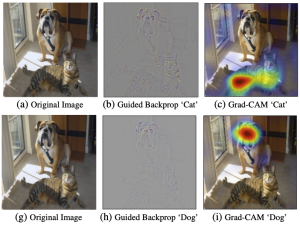
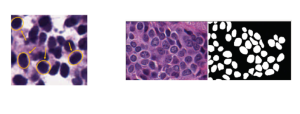
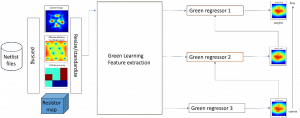
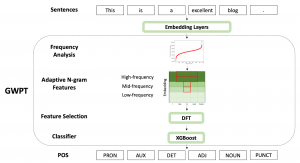
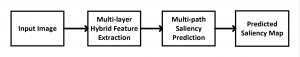
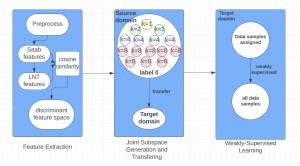
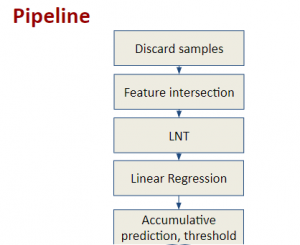
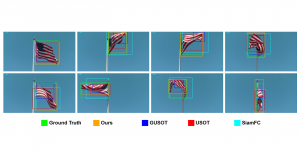
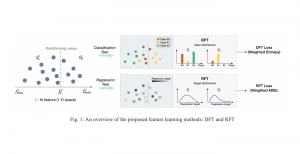
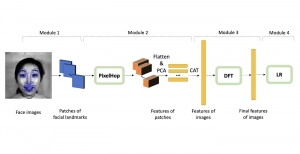
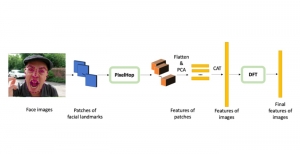
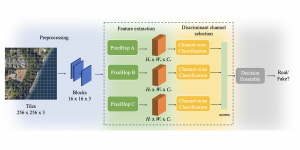
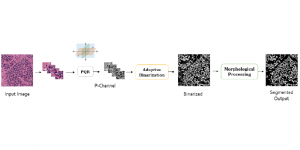
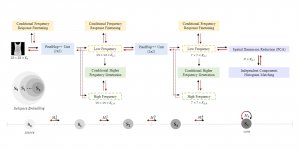
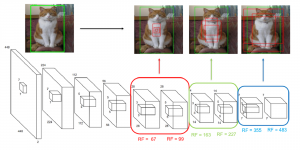
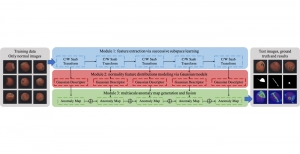
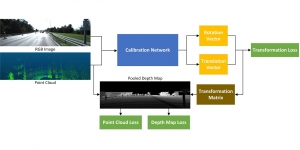
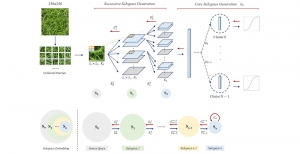
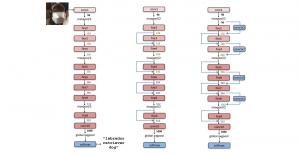
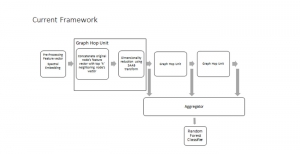
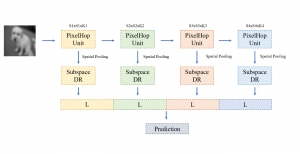
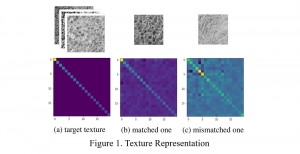
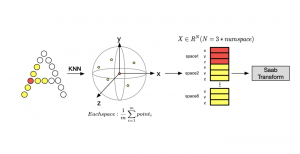
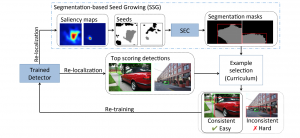
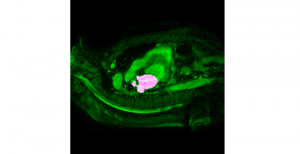
The tumor microenvironment includes many types of cells around the tumor. Doctors often assess it using biomarkers like PD-L1 or CD68. To measure how many cells express these markers—called positive cells—we use image analysis and machine learning models to identify positive cells and compute their count and ratio. Usually, immunohistochemistry (IHC) images are used as the input of the model because they are relatively low-cost, while multiplex immunofluorescence (IF) images are used as ground truth due to their high accuracy. A major research goal is to predict positive cell locations from IHC images.
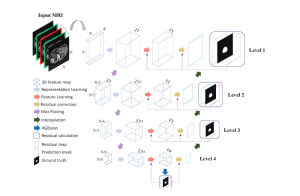
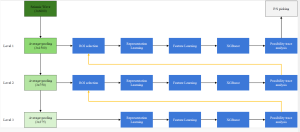
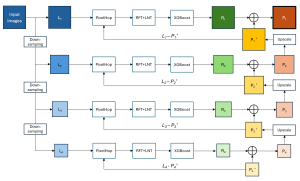
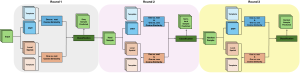
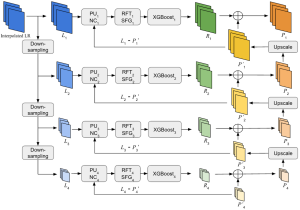
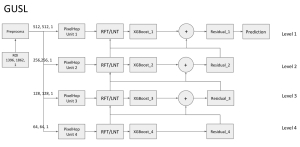
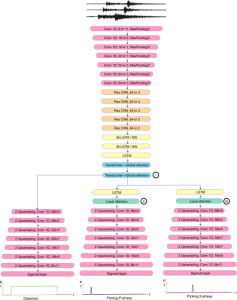
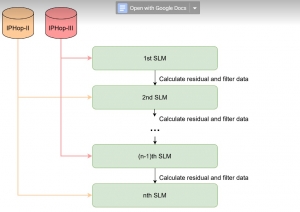
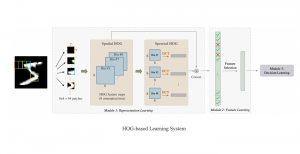
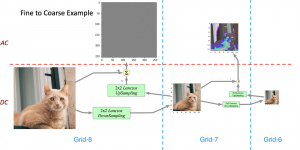
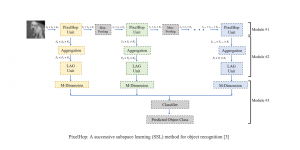
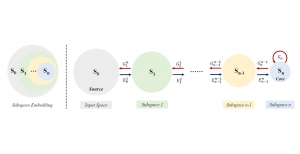
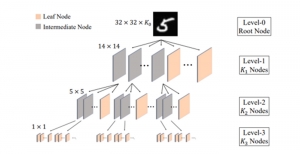
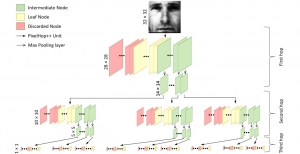
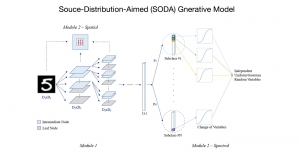
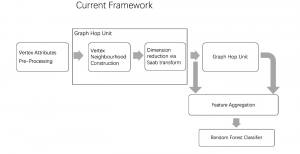
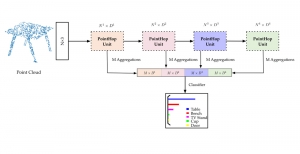
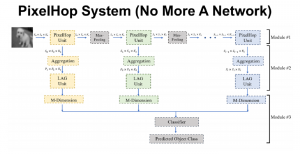
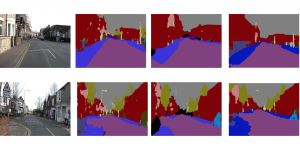
Our current work explores the use of the Green U-shaped Learning (GUSL) pipeline to align input IHC images with the ground truth IF images. GUSL is well-suited for this task because it enables pixel-wise prediction from coarse to fine resolution. It can detect positive cells at a coarse level and progressively refine predictions. GUSL has also shown strong performance in related tasks like kidney segmentation.
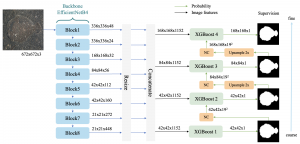
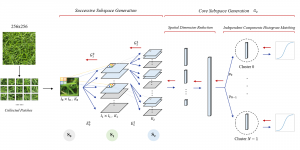
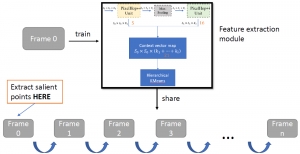
Another approach we explore is using GUSL to generate predictions on other medical images, such as H&E-stained images, DAPI, and LAP2. By producing segmentation results from H&E, DAPI, LAP2, and IHC inputs, and then taking a weighted average, we aim to further improve prediction accuracy.
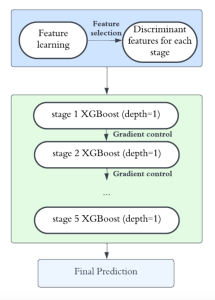
Currently, many machine learning methods have been developed to solve this problem, but they often suffer from high computational cost (FLOPs) and large model size. In addition, the limited size of medical datasets presents another major challenge. Green learning offers a promising solution to these issues and contributes to noninvasive biomarker prediction in this research field, helping reduce the need for expensive and labor-intensive staining methods.